ANTINATURAL SELECTION
BACTERIAL ENCRYPTION
What makes us human? This is one of the initial questions addressed in this project, but we think it cannot be detached from the notion of identity. Since the discovery and study of DNA, perception of humanity and our uniqueness as individuals has gradually changed overtime. And even though we share up to 90% of our code with other mammals (like mice), and more than 99% among humans, the sequences in our genome that make us different are being thoroughly studied in order to unequivocally identify us. This has been of concern for artists and activists when biological information such as DNA or biometric data are used as a means of control and incriminating evidence by certain governments. On the other hand, from a transhumanist perspective, the complete understanding of the human genome would suppose its modification, thus managing to eradicate the disease, and eventually slow down aging.
Both approaches have and will have much greater ethical, social and environmental implications than those we already face today. Through this project we propose, on the one hand, the appropriation of our DNA information for its reinterpretation through other species, which also implies a form of relationship with other living beings, as well as an evolution of our identity outside of the human body. Subsequently, mutations in our DNA inside other species will be recovered and used as a form of encryption for digital systems that regularly store and recognize individuals through their biological data (for instance facial, finger, iris, voice recognition), this as a speculative form of defence against such control, while also allowing us to observe 'human identity' modified by the evolutionary processes of other species.
For the first stage of the project, extensive research was carried out on the sequences of our DNA that could give clues of who we are or what makes us human. We selected 6 transcendent genes and sequences:
Sequence 1. HAR1 region (Human Accelerated Region 2). It is a non-coding region that has been linked to the evolution of the human brain. It is conserved in all vertebrates but is particularly different in humans, and it is the HAR region that shows the greatest number of differences from the chimpanzee. The oligos for the amplification of the HAR1 region were specifically designed for this work. The sequences are DMGLHar1F 3 'TGCACGTGAGTTAGAG 5' and DMGLHarR 3 'GCCTTCCTCCTCTTG 5'.
Sequence 2. Gene that codes for Amelogenin. It is a protein that is produced in the development of tooth enamel. It is a single copy gene found on the sex chromosomes. It is used in forensic genetics as one of the parameters for the determination of chromosomal sex.
The oligos used were AmelF 5 & # 39; -CTGATGGTTGGCCTCAAGCCTGTG -3 & # 39; and AmelR 5 & # 39; -TAAAGAGATTCATTAACTTGACTG-3 & # 39. The PCR product size is a single 977 bp band for a person with XX chromosomes and two bands, one 977 bp and one 788 bp for a person with XY chromosomes.
Sequence 3. Universal amplification region for the identification of animal species for forensic application. This sequence corresponds to a region corresponding to the cytochrome b sequence, and was determined by S. K. Verma and L. Singh. The sequences of the corresponding oligos are mcb398 5'-TACCATGAGGACAAATATCATTCTG-3 'and mcb869 5'-CCTCCTAGTTTGTTAGGGATTGATCG-3 '. These oligos have been used successfully for the determination of the origin of animal and human meat.
Sequence 4. The HACNS1 sequence is located in an intron of the CENTG2 gene, also known as HAR. It is hypothesized that this region is part of a repressor involved in the expression of genes related to the formation of the hand, ankle and thumb. In particular, the evolution of this sequence is considered to have contributed to the development of opposable thumbs and possibly the development of the ability to walk on two legs. Among the regulatory regions of genes found in the human genome, HACNS1 is the one that has had the most changes in human evolution since its evolutionary line separated from the chimpanzee. The oligos to amplify this region were designed specifically for this work, with sequences DMGLHar2F 3 'CATACAGTAGCTTCATTG 5' and DMGLHar2R 3 'CAAGTGTGGCAAAAATGAC 5' (the latter partially modified from Kenta Sumiyama and Naruya Saitou, 2011).
Sequence 5. Gene that codes for the receptor on CD4 T lymphocytes. This gene is used as an identity marker in forensic genetics. The oligos for the amplification of this gene were obtained from the Short Tandem Repeats (STR) database. The CD4 gene encodes a glycoprotein membrane that is related to the immune response in T cells. This receptor is the entry pathway for the human immunodeficiency virus (HIV). The oligos used were AmelF 5 & # 39; - TTGGAGTCGCAAGCTGAACTAGC -3 & # 39; and AmelR 5 & # 39; -GCCTGAGTGACAGAGTGAGAACC -3 & # 39. The size of the product varies according to the individual alleles of the identified person. The gene is located on chromosome 12 in human2.
Sequence 6. D18S51 region. This region was implemented as a marker for the identification of corpses in Mexico, specifically in the Veracruz area. The need to design methodologies that allow to identifying human corpses arises from the problems generated by organized crime and its victims. The oligos to amplify this region are: D18S51F 5 & # 39; -TTCTTGAGCCCAGAAGGTTA-3 & # 39; and D18S51R 5 & # 39; -ATTCTACCAGCAACAACACAAATAAAC-3 & # 39 ;. The region amplified by these oligos varies in size according to individual identity and is approximately 300 to 400 bp.

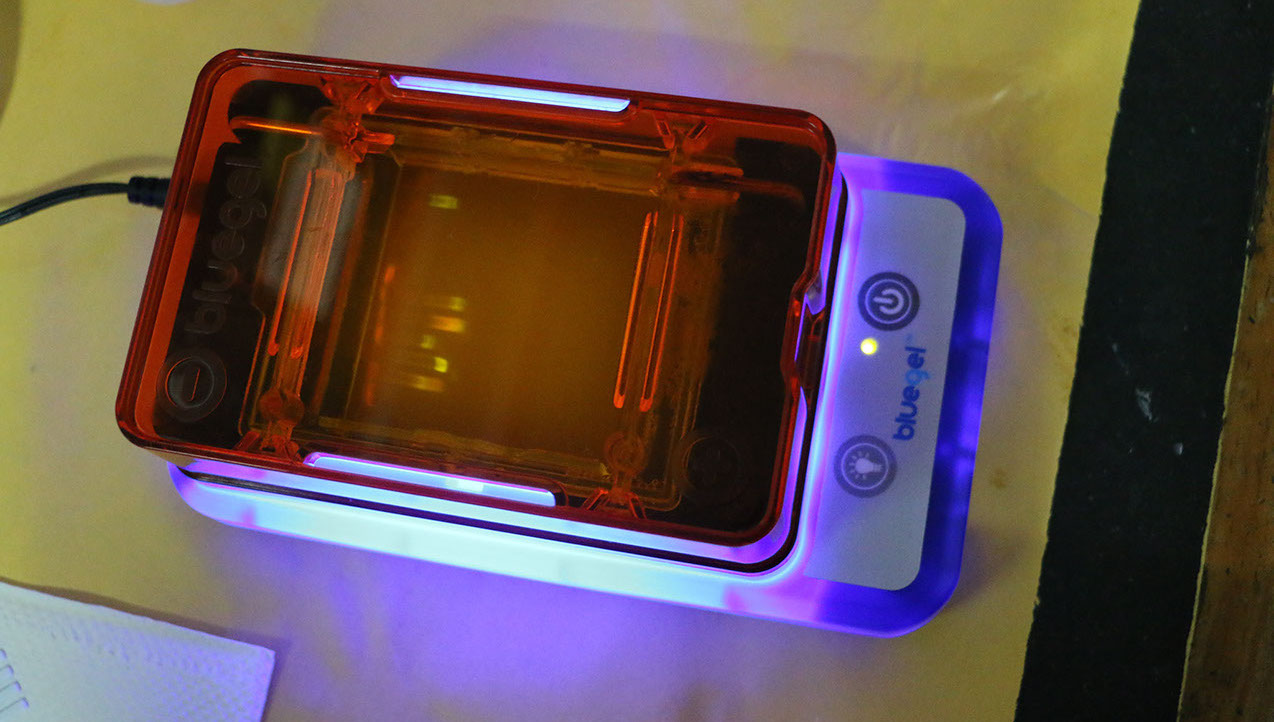

We extracted DNA from our blood and amplified the aforementioned regions by PCR. The amplification conditions were calculated with the Vector NTI v.6 program and the amplification was carried out in a mini16 thermal cycler (mini PCR M.R), with 16 wells. The sequences were added to different plasmids and later inserted into DH5alpha bacteria (Escherichia coli). For the growth of the bacteria, 3D-printed stamps with the shape of the artist's fingerprint and iris were used. The intention of making them grow in such patterns is that the bacteria would modify not only the DNA sequences inserted in their plasmids (mutations caused by UV rays), but that at the same time they would modify the forms of the biometric data of the artist. In order to recognize which bacteria inherited the plasmids with DNA when reproducing, the white / blue protocol was used, in which the vectors (the inserted plasmids) contain a selective marker. In this case bacteria that transmit the artist's DNA appeared in white, while a few that did not succeed were coloured blue.
After several generations of bacterial growth, the sequences of their plasmids were recovered and they are currently being sequenced to use the mutation factor as material for digital artworks, in which the interpretation of identity will be made from these mutations. It is in our interest to repeat this process in other types of beings, for example plants.


